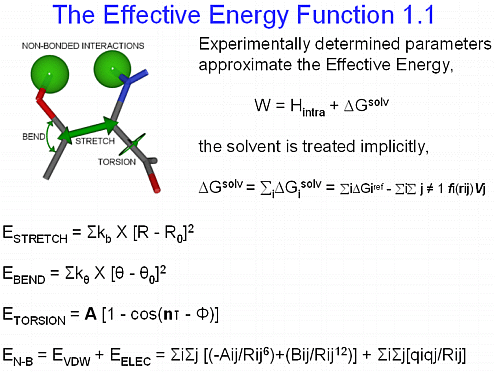
The relative stability of a structure is related to the free energy change,
where ΔW is the effective energy change and TΔS is the configurational entropy change. We approximate the free energy of coiled coil models using molecular dynamics simulations with the effective energy function EEF1.1 [Lazaridis, T., Karplus, M. (1999), Proteins 35:133-152]. The entropic components are computed using standard thermodynamic equations coded in FORTRAN. For example, the side chain entropy is computed from the probability distribution of each side chain torsional angle, obtained by rotating each of them independently of the others about their heavy atom (C,N,O, and S) χ bonds,
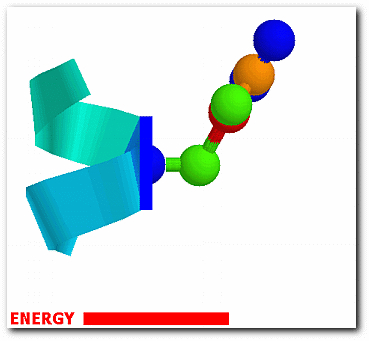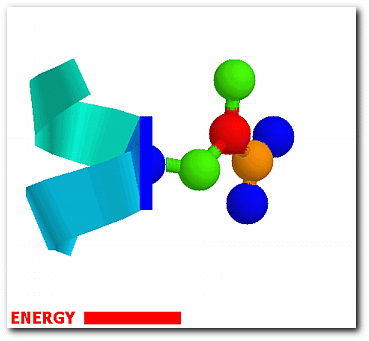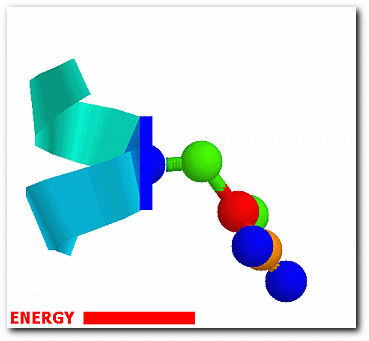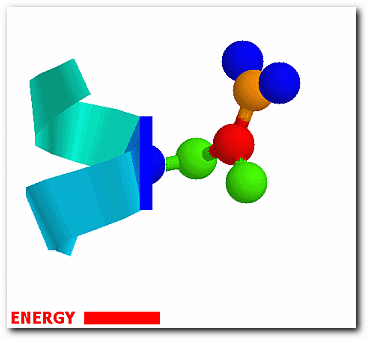
The probability of each conformation from the effective energy profiles is,
exp[W(ω)/RT] p(ω) = ------------------- ∫exp[W(ω)/RT]dω
where ω denotes one of the sampled side-chain conformations and W(ω) is the associated energy. The conformational entropy of a structure is,
Sconf = -RΣN(χ)[∫p(ω)ln p(ω)dω]
where the outer summation is over the total number N(χ) of torsional angles in the protein.
Our free energy calculation protocol using a conformational search can discriminate the correct oligomeric state of homotypic parallel coiled coils and reproduce some generally accepted rules. The same method has also been employed to investigate the tendency of certain sequences to form antiparallel coiled coil dimers, trimers, and tetramers.

Jorge Ramos, Ph.D.
-|- SEMPER FI -|-
CURRENT DUTIES/HOBBIES
CONTACT INFORMATION
Department of Chemistry
Queens College of New York
65-30 Kissena Blvd.
Queens, NY 11367
tel: +1-718-997-3273
EMAIL:jr_starwind@netzero.net
